
- #How to use restriction enzymes snapgene viewer update#
- #How to use restriction enzymes snapgene viewer software#
Give you easy ways to troubleshoot these situations.ĭNA Fragment with Sticky Ends or Blunt Ends Into restriction enzyme based cloning, we’ll tackle some of the challenges, and Therefore, in this article, we’ll look deeper Sometimes you justĬan’t get those elusive white colonies on your plate, even after following the But, performing a DNA cloningĮxperiment using restriction enzymes is not as easy as it seems. Restriction enzymes in your DNA cloning experiment. Most of the time, as a researcher, you can’t avoid using Restriction enzymes are enzymes that cut DNA at or near a specific target sequence, called a restriction site. Also referral discount codes have been discontinued (hence the strikethrough text above.Since the early 1970s, restriction enzymes have become an important part of cloning and many other applications, including DNA mapping.
#How to use restriction enzymes snapgene viewer update#
UPDATE (): SnapGene is changing their licensing scheme (no permanent licenses anymore, only yearly subscription fees). If you are considering to purchase a license, you get a 10% discount when using my email address ( as a discount code.

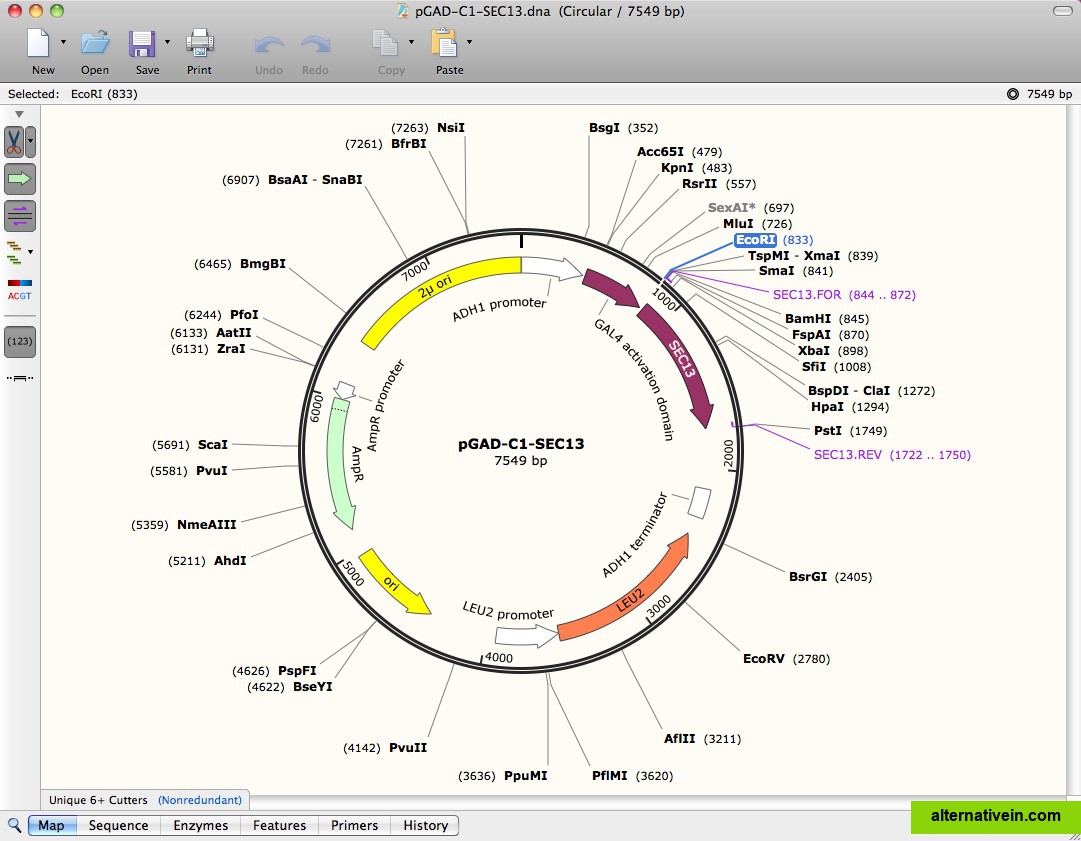
Among them is Addgene, a very useful nonprofit distribution channel for DNA constructs (. Many companies and labs have started to use SnapGene (many of them based on our recommendations). For example, if you want to isolate an insert that can be excised only with a restriction enzyme that cuts also within your insert as shown in the image: Despite two additional BssHII cuts within the sequence, it is possible to isolate the 1985-bp-long BssHII-SalI fragment. This is exactly the type of work that computers are ideal for! Sometimes, you also want to do a partial digest on purpose. two enzymes, which both cut your DNA at multiple sites. However, this requires manually calculating all possible fragment lengths, which can be quite some work if you have used e.g. Then you get typically a complex pattern, from which you very often can deduce (with some effort), whether your construct is OK or not. This comes in handy when you do an analytical digest and you did not add enough enzyme. However, Snapgene has still not implemented a feature that we would like to have: partial restriction enzyme digests.


The program is able to read all files within that folder and you can perform actions on the collection (e.g.
#How to use restriction enzymes snapgene viewer software#
Since I last wrote about the SnapGene software ( ), many good things have happened: Snapgene is a software for the wet lab molecular biologist, who does lots of cloning work (construct design and annotation).


 0 kommentar(er)
0 kommentar(er)
